Comparing In Vitro and In Vivo Models as Part of Pre-Clinical Studies for COVID-19 Medicines
Poppy Bradley1
1University of Bath, Department of Natural Sciences
Youth STEM Matters, no. 1 (2021); https://doi.org/10.51892/ysm.1.202103P. Bradley, "Comparing In Vitro and In Vivo Models as Part of Pre-Clinical Studies for COVID-19 Medicines," Youth STEM Matters,
no. 1, 2021.
If you cite this article or use it in any academic activities please let us know by e-mailing us at editor@youthstem2030.org.
Abstract
Introduction
Pre-clinical studies play a key part in the drug discovery pathway. Following non-clinical studies, pre-clinical studies may be performed to address the pharmacological profile of any promising lead candidates. This encompasses a wide variety of parameters requiring consideration, including the drug’s pharmacokinetics, pharmacodynamics, efficacy and toxicological properties [1]. Cell and animal models are used to determine administration and off-target interactions, ensuring the drug is fit-for-purpose [2]. Approximately 95% of compounds are withdrawn due to safety issues or a lack of efficacy [3]. Thus, there is a disparity in the translation of medicines from “bench to bedside.” Development and improvement of pre-clinical studies should permit the optimisation of compounds for the clinic while maintaining desirable pharmacological profiles [4].
Translational studies have been used to model disease and have become increasingly vital in the study of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) during the COVID-19 pandemic. COVID-19 symptoms are characterised by a high temperature, a continuous cough, and a change or loss of the senses of smell and taste [5]. Upon infection, SARS-CoV-2 binds to the host cell receptor angiotensin-converting enzyme 2 (ACE2) facilitating viral loading. These ACE2 receptors are primarily expressed in the airway epithelia and vascular endothelium, which upon infection presents a range of respiratory symptoms and can progressively develop to severe pneumonia [6]. Therefore, to study the virus in both kinds of models, they must be able to express the ACE2 receptor to allow viral infection. Whilst in vitro and in vivo studies have their strengths and weaknesses in developing drugs against SARS-CoV-2, by accumulating data from both types of studies, drug candidates can advance further. The close links and interdependence between these studies also highlight the importance of research into more robust translational science.
In Vitro Approaches
In vitro studies that develop a pharmacological profile for a drug candidate typically follow hypothesis-driven work utilising cell lines or intact tissue. This includes screening for its affinity and selectivity for a target, as well as its functional efficacy [7]. In terms of the drug discovery process, these studies can mimic in vivo studies at a lower expense [8], whilst reducing the risk of clinical failure [9]. For pharmaceutical companies, this is where most funding is allocated. It is key to ensure a drug is worth taking forward from a financial standpoint. Drugs are initially tested in several different types of model mammalian cell lines which can be cultured and divide indefinitely. Cells can be manipulated to model disease and help test drugs without having to put them straight into animals. Vero-E6 cell lines were used for preclinical studies after the 2002 SARS outbreak [10]. These cells come from African green monkey kidneys and were previously used as models for influenza infection. This is due to their ability to retain a high viral load, which is important for pre-clinical studies as the cellular models will be able to retain the virus, which can then be treated with different drugs. This model has been widely used because of the similarity between the receptor site of SARS-CoV-2 and SARS in its receptor site, angiotensin-converting enzyme 2 (ACE2) [11]. This allows, as discussed earlier, for viral entry and replication in the model. It is now well-established as an in vitro model for COVID-19 [12]. The other approach taken is using intact tissue taken directly from patients or animals with the disease [13]. This is crucial when it comes to lightly modelling as disease-specific changes to the tissues or cells can be monitored. Therefore, it is clear that in vitro approaches can mimic the effect of COVID-19 on cells. This has already been demonstrated in many studies to test different drugs, including remdesivir and hydroxychloroquine.
Remdesivir and Hydroxychloroquine
In vitro methods come with their advantages and limitations, as demonstrated by pre-clinical studies of remdesivir and hydroxychloroquine (HCQ) for COVID-19 treatment. Remdesivir, which was originally developed to treat Ebola, was examined as a candidate to treat COVID-19. Remdesivir works by inhibiting RNA-dependent RNA Polymerase (RdRp), the viral polymerase enzyme responsible for viral replication [14]. Following pre-clinical studies, it was shown that it was effective in reducing COVID-19 viral loads in cells. In Vero-E6 cells, remdesivir was measured to have an EC50 (i.e., the concentration of the drug required to illicit 50% of a maximal response) of 0.77 µM [15]. Singh and colleagues examined other pre-clinical studies using other drugs, including remdesivir and HCQ. They concluded that remdesivir had the optimal safety profile over the other drugs, yet more clinical work was required to make a judgement [16].
Regarding HCQ, it is generally used as an antimalarial. In pre-clinical studies utilising Vero-E6 cells, HCQ was found to be a more potent inhibitor of SARS-CoV-2 with an EC50 of 4.51 µM [17]. However, alongside this, it was found to be more cytotoxic than the drug it was being compared to, chloroquine (another antimalarial) [18]. In addition, its mechanism of action against SARS-CoV-2 is still unknown [19].
Considering the EC50s of both drugs on the same cellular model, both have very similar efficacy. Ultimately, remdesivir was approved for COVID-19 treatment across the world following success in the clinic [20]. On the other hand, emergency market authorisations were removed for HCQ as a treatment for COVID-19 due to a lack of efficacy [21]. This demonstrates a further need for thorough pre-clinical work, given that the efficacy data was so similar [22]. This could suggest that using just Vero-E6 cells does not translate wholly to the clinic and that the cultures may not fully represent how different human tissues interact with the virus and the drug, rather than just one isolated tissue. The same assay, CCK-8 assays, were also used in each case. More information from different assays in how both drugs work may well have been uncovered had different cellular models been used.
The predominant advantage of using in vitro models is that a specific cell type can be targeted, with a choice of level of receptor expression. Models have become more complex and physiologically relevant. In COVID-19 studies, a culture system called human airway epithelium air-liquid interface (HAE-ALI) has helped to test the efficacy of drugs such as remdesivir and HCQ. It reflects the stratified columnar organisation of the human airways, having an apical face that meets the “airway.” Reflecting on the polarity of cells has allowed for a valid model to test hypotheses of HCQ entry [8].
Whole-tissue approaches allow signalling pathways to be maintained with a physiological representation of receptor expression. Organoids have addressed the need for accurate in vivo models, as well as permitting the avoidance of the use of whole animals [23]. Not only this, organoids grown from cell lines to form full lung-like tissue accurately reflected the “cytokine storm” seen in severely ill COVID-19 patients; a clinical symptom that has been hard to model and test with HCQ [24]. They also highlighted signalling mechanisms by which this immune response works, lending knowledge to target identification [25].
However, there is still a need for translatable models. Many tissues do not accurately reflect the physiology of human tissue, especially in cell lines [26]. Induced pluripotent stem cells lack scalability and their nature means it is hard to have a structurally integral model in mass screens. Primary cells still require animal tissue, which only remains viable in the short term [27]. In terms of the drug discovery process, in vitro approaches are largely moving towards 3D models to help minimise costs. In a time where solutions are required rapidly, it may mean different approaches are needed.
In Vivo Approaches
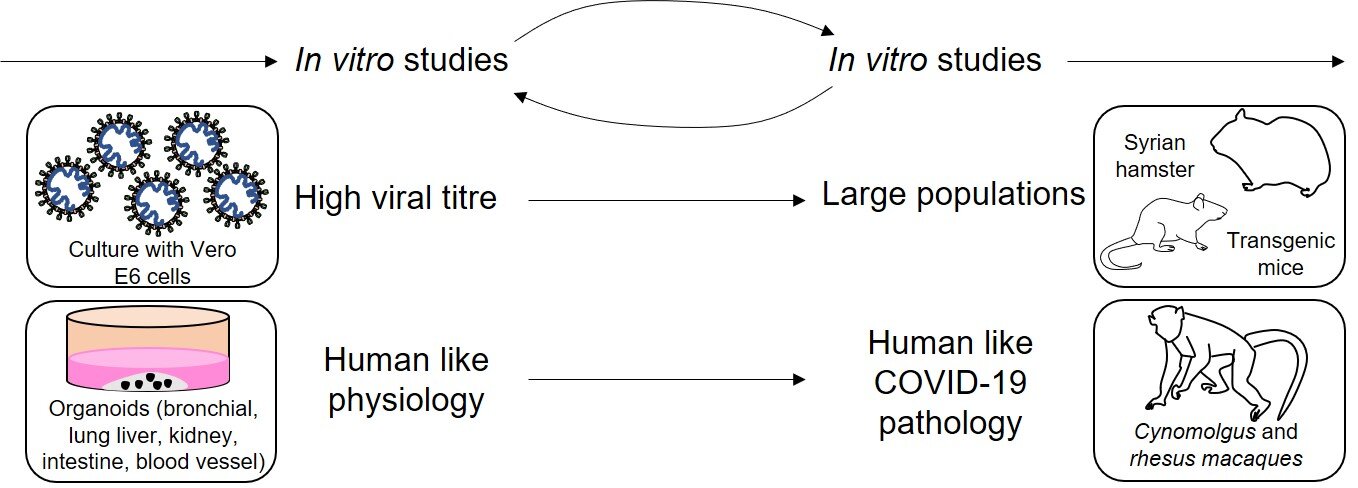
The next stage of pre-clinical study is needed to gain an idea of how the drug interacts within whole animals, being the first point at which the drug has been administered into a whole organism. They act as a predictive link to the clinic, quantifying the efficacy of the drug. It also gives an indication of any unpredicted side effects. Finally, they give an idea of the drug’s translatability into clinical studies [1]. Typically, one small mammal (such as a rodent) and one large mammal (usually a non-human primate (NHP)) are used, dependent on the trial’s needs. These choices are represented by the decision flowchart shown in Fig. 1.
Figure 1 - Decision flowchart showing some of the models that can be used to model SARS-CoV-2 pathophysiology according to specific needs of the study. In vitro studies inform in vivo studies and vice versa. By using cells that can generate a high viral titre, such as Vero-E6, the information can be translated into larger in vivo models, such as hamsters and mice, which can acquire COVID-19 infection. Human organoids can mimic certain aspects of COVID-19 pathophysiology, and NHPs like macaques scale this up further. This shows how aspects of in vitro study can then inform in vivo studies further down the pipeline. Other non-specific in vitro models include airway epithelial cells and cell lines such as Caco-2, Calu-3, HEK293T and HuH7. Non-specific in vivo models include ferrets and cats. Figure constructed by Author, based on Ref [26].
In vivo studies come with both advantages and limitations. The main advantage is that many of the NHPs have a near-full biochemical likeness to humans. There is strong receptor homology between humans and rhesus and cynomolgus macaques. NHP models mimic human disease so accurately that even the epithelial necrosis commonly seen with severely ill patients can be measured as an outcome in NHP trials. Ferrets have also acted as a good model for mild to moderate disease [28]. Using these findings combined with imaging has allowed researchers to gain an idea of how the disease may present in clinical settings [29]. Furthermore, transgenic mice susceptible to chronic obstructive pulmonary disease (COPD) have been produced, so work can be done to improve mortality for those with complex health needs. COPD is characterised by breathlessness, a chesty cough, and persistent wheezing, like some of the symptoms of COVID-19. Having models like this are important for having the capability to model complex disease states [30]. In vivo models have also shown severe symptoms, such as cytokine release syndrome – which is extremely difficult to model in cells [31]. Further improvements have been made to the validity of in vivo models. Genetic modifications to mice have meant that they present humanised ACE2 receptors, making them susceptible to infection. Another approach is making a murine SARS-CoV-2 virus, making virions able to enter cells through ACE2 receptors [32]. This has helped to generate more accurate “human” models.
However, there are drawbacks associated with such studies. There are three types of validity to consider: isomorphic, homologous, and predictive validity. Isomorphic validity deals with whether the model reflects the pathological changes associated with human disease. Not only do pharmaceutical companies have competing interests, but studies are also done on a small scale with strictly defined readouts. There have already been instances where genetically modified mouse strains were incorrectly described in literature, making comparisons difficult [30]. In one case, the adeno-associated virus delivery system created to infect mouse cells presenting ACE2 was not specific enough to only infect cell types permissive to SARS-CoV-2 in humans. This meant that the data collected was hard to interpret in the context of human airway infection. As well as this, many of these studies were carried out using intestinal, kidney, and lung cell lines. This means the models may have had a lack of immune components (for example macrophages and natural killer cells) which are essential for the body’s modulation of COVID-19. More appropriate cell models with this consideration include both upper and lower airway organoid models of SARS-CoV-2 infection [33].
Homologous validity considers how much of the aetiology of human disease is reflected. There is said to be no ‘good’ animal model at this point that fully shows a human-like COVID-19 viral infection in its tissues. Non-engineered mice are innately resistant to infection, due to the incompatibility of S protein with murine ACE2 receptors. This complicates model development [32]. ACE2 transgenic mice show no severe clinical symptoms, and instead, have a 40% mortality due to brain tropisms [28]. These problems only hinder drug translation further.
Predictive validity considers whether the model is translatable. This is the extent to which how accurate the model is compared to the tissue being studied. Hamsters have been used due to their strong homologous validity of ACE2 receptors. However, there are limited reagents that can be used alongside a lack of genetic traceability. Combined with a lack of widespread availability, this makes the model hard to scale [30]. NHPs present a problem in terms of the ethics and large costs associated. Limited availability is an issue in a time where most research institutions are conducting NHP studies [34]. In terms of the drug discovery pathway, both types of studies are required to gather the required data. Where in vivo studies are more intensive and in the context of the whole animal, they do yield important results and give us the best indicator of how a drug will be tolerated. Further optimisation of models is required to enhance this further.
Conclusion
To conclude, by considering models of disease in two different settings, it is clear to see that both approaches come with their own advantages and limitations. Whilst in vitro models provide a good knowledge of a pharmacological profile, it is hard to see wider effects and to construct more complex models. As much as in vivo studies act as a predictive link to the clinic, many models are lacking in key elements of validity; this prevents researchers from gaining a full perspective for drug action. As for the wider implications of this, it highlights the importance of using both types of study together to tailor to each compound. It is clear from the examples, such as remdesivir, pre-clinical studies are effective in identifying effective compounds for medical treatment. Without doing these studies there would be no evidence to move into the clinic, these studies can also reveal important evidence to repurpose in earlier stages of the pathway. This is through the close morphology and molecular physiology of the models used to host the COVID-19 virus. It also highlights a need for investment into translational science. This will allow the optimisation of pre-clinical studies to make compounds safer, more efficacious, and cost-effective. Both approaches are essential to the drug discovery process and only with the right investment can these models be made appropriate for their course of study. In a time where there is such a great need, it emphasises the importance of both pre-clinical studies and drug development to our world.
Acknowledgements
I would like to thank my Mum and Dad for always believing in me not only as a scientist, but also as a person.
Conflict of Interest Statement
Poppy Bradley is, at the time of publication, the Coordinating Managing Editor for the Youth STEM Matters journal. The double-blind editorial process has, however, ensured that the Reviewers were unaware of this fact. No other conflicts of interest are declared.
Author's Notes
All figures, images, or tables were created by the author, unless otherwise mentioned in the description provided for said figure.
References
[1] R. G. Hill, Drug discovery and development: technology in transition. Churchill Livingstone, 2012.
[2] A. G. Polson and R. N. Fuji, "The successes and limitations of preclinical studies in predicting the pharmacodynamics and safety of cell-surface-targeted biological agents in patients," British Journal of Pharmacology, vol. 166, no. 5, pp. 1600-1602, 2012. Available: https://doi.org/10.1111/j.1476-5381.2012.01916.x
[3] T. Hartung, "Look back in anger - what clinical studies tell us about preclinical work," ALTEX, vol. 30, no. 3, pp. 275-291, 2013. Available: https://doi.org/10.14573/altex.2013.3.275.
[4] A. A. Seyhan, "Lost in translation: the valley of death across preclinical and clinical divide – identification of problems and overcoming obstacles," Translational Medicine Communications, vol. 4, no. 1, p. 1-9, 2019 Available: https://doi.org/10.1186/s41231-019-0050-7.
[5] C. Menni et al., "Real-time tracking of self-reported symptoms to predict potential COVID-19," Nature Medicine, vol. 26, no. 7, pp. 1037-1040, 2020. Available: https://doi.org/10.1038/s41591-020-0916-2.
[6] A. G. Harrison, T. Lin, and P. Wang, "Mechanisms of SARS-CoV-2 Transmission and Pathogenesis," Trends Immunol, vol. 41, no. 12, pp. 1100-15, Dec 2020. Available: https://doi.org/10.1016/j.it.2020.10.004.
[7] H. G. Vogel, W. H. Vogel, B. A. Schölkens, J. Sandow, G. Müller, and W. F. Vogel, Drug Discovery and Evaluation: Pharmacological Assays Second Completely Revised, Updated, and Enlarged Edition, 2nd ed. Berlin, Heidelberg: Springer, 2002.
[8] S. Hao, K. Ning, C. A. Kuz, K. Vorhies, Z. Yan, and J. Qiu, "Long-Term Modeling of SARS-CoV-2 Infection of In Vitro Cultured Polarized Human Airway Epithelium," MBio, vol. 11, no. 6, pp. e02852-20, 2020. Available: https://doi.org/10.1128/mBio.02852-20.
[9] A. K. Singh, G. Mishra, A. Maurya, G. T. Kulkarni, and R. Awasthi, "Biofabrication: An interesting tool to create in vitro model for COVID-19 drug targets," Medical hypotheses, vol. 144, pp. 110059-110059, 2020. Available: https://doi.org/10.1016/j.mehy.2020.110059.
[10] M. Kaye, et al.,"SARS-associated coronavirus replication in cell lines," (in eng), Emerging infectious diseases, vol. 12, no. 1, pp. 128-133, 2006. Available: https://doi.org/10.3201/eid1201.050496.
[11] S. Weston et al., "Broad Anti-coronavirus Activity of Food and Drug Administration-Approved Drugs against SARS-CoV-2 In Vitro and SARS-CoV In Vivo," Journal of virology, vol. 94, no. 21, pp. e01218-20, 2020. Available: https://doi.org/ 10.1128/JVI.01218-20.
[12] C. Woolsey et al., "Establishment of an African green monkey model for COVID-19 and protection against re-infection," Nature Immunology, vol. 22, no. 1, pp. 86-98, 2021. Available: https://doi.org/10.1038/s41590-020-00835-8.
[13] T. P. Sheahan et al., "An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice," Science Translational Medicine, vol. 12, no. 541, 2020. Available: https://doi.org/10.1126/scitranslmed.abb5883.
[14] A. Frediansyah, F. Nainu, K. Dhama, M. Mudatsir, and H. Harapan, "Remdesivir and its antiviral activity against COVID-19: A systematic review," Clinical Epidemiology and Global Health, vol. 9, pp. 123-127, 2021. Available: https://doi.org/10.1016/j.cegh.2020.07.011.
[15] M. Wang et al., "Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro," Cell Research, vol. 30, no. 3, pp. 269-271, 2020. Available: https://doi.org/10.1038/s41422-020-0282-0.
[16] A. K. Singh, A. Singh, R. Singh, and A. Misra, "Remdesivir in COVID-19: A critical review of pharmacology, pre-clinical and clinical studies," Diabetes Metabolic Syndrome, vol. 14, no. 4, pp. 641-648, 2020. Available: https://doi.org/10.1016/j.dsx.2020.05.018.
[17] J. Liu, R. Cao, and M. Xu, "Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro," Cell Discovery, vol. 6, no. 1, p. 16, 2020. Available: https://doi.org/10.1038/s41421-020-0156-0.
[18] X. Yao et al., "In Vitro Antiviral Activity and Projection of Optimized Dosing Design of Hydroxychloroquine for the Treatment of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2)," Clinical Infectious Diseases, vol. 71, no. 15, pp. 732-9, 2020 Available: https://doi.org/10.1093/cid/ciaa237.
[19] D. Maciorowski et al., "A Review of the Preclinical and Clinical Efficacy of Remdesivir, Hydroxychloroquine, and Lopinavir-Ritonavir Treatments against COVID-19," SLAS DISCOVERY: Advancing the Science of Drug Discovery, vol. 25, no. 10, pp. 1108-1122, 2020. Available: https://doi.org/10.1177/2472555220958385.
[20] Y. Wang et al., "Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial," The Lancet, vol. 395, no. 10236, pp. 1569-1578, 2020. Available: https://doi.org/10.1016/S0140-6736(20)31022-9.
[21] B. Singh, H. Ryan, T. Kredo, M. Chaplin, and T. Fletcher, "Chloroquine or hydroxychloroquine for prevention and treatment of COVID‐19," Cochrane Database of Systematic Reviews, no. 4, 2021. Available: https://doi.org/ 10.1002/14651858.CD013587.pub2.
[22] R. E. Ferner and J. K. Aronson, "Chloroquine and hydroxychloroquine in covid-19," BMJ, vol. 369, 2020. Available: https://doi.org/10.1136/bmj.m1432.
[23] H. Clevers, "COVID-19: organoids go viral," Nature Reviews Molecular Cell Biology, vol. 21, no. 7, pp. 355-6, 2020. Available: https://doi.org/10.1038/s41580-020-0258-4.
[24] C. Tindle et al., "Adult Stem Cell-derived Complete Lung Organoid Models Emulate Lung Disease in COVID-19," bioRxiv, 2020. Available: https://doi.org/ 10.1101/2020.10.17.344002.
[25] J. Huang et al., "SARS-CoV-2 Infection of Pluripotent Stem Cell-Derived Human Lung Alveolar Type 2 Cells Elicits a Rapid Epithelial-Intrinsic Inflammatory Response," Cell Stem Cell, vol. 27, no. 6, pp. 962-73,2020. Available: https://doi.org/10.1016/j.stem.2020.09.013.
[26] K. Takayama, "In Vitro and Animal Models for SARS-CoV-2 research," Trends in Pharmacological Sciences, vol. 41, no. 8, pp. 513-517, 2020/08/01/ 2020, Available: https://doi.org/10.1016/j.tips.2020.05.005.
[27] A. M. Tatara, "Role of Tissue Engineering in COVID-19 and Future Viral Outbreaks," Tissue Engineering Part A, vol. 26, no. 9-10, pp. 468-74, 2020. Available: https://doi.org/10.1089/ten.tea.2020.0094.
[28] S. N. Neerukonda and U. Katneni, "A Review on SARS-CoV-2 Virology, Pathophysiology, Animal Models, and Anti-Viral Interventions," Pathogens (Basel, Switzerland), vol. 9, no. 6, p. 426, 2020. Available: https://doi.org/10.3390/pathogens9060426.
[29] P. Maisonnasse et al., "Hydroxychloroquine use against SARS-CoV-2 infection in non-human primates," Nature, vol. 585, no. 7826, pp. 584-7, 2020. Available: https://doi.org/ 10.1038/s41586-020-2558-4.
[30] M. D. Johansen et al., "Animal and translational models of SARS-CoV-2 infection and COVID-19," Mucosal Immunology, vol. 13, no. 6, pp. 877-91, 2020. Available: https://doi.org/10.1038/s41385-020-00340-z.
[31] J. Wang et al., "The ACE2-deficient mouse: A model for a cytokine storm-driven inflammation," (in eng), FASEB journal: official publication of the Federation of American Societies for Experimental Biology, vol. 34, no. 8, pp. 10505-10515, 2020. Available: https://doi.org/10.1096/fj.202001020R.
[32] K. H. Dinnon et al., "A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures," Nature, vol. 586, no. 7830, pp. 560-566, 2020. Available: https://doi.org/10.1038/s41586-020-2708-8.
[33] S. R. Leist, A. Schäfer, and D. R. Martinez, "Cell and animal models of SARS-CoV-2 pathogenesis and immunity," Disease Models & Mechanisms, vol. 13, no. 9, 2020. Available: https://doi.org/10.1242/dmm.046581.
[34] S. G. P. Funnell et al., "Emerging preclinical evidence does not support broad use of hydroxychloroquine in COVID-19 patients," Nature Communications, vol. 11, no. 1, p. 4253, 2020. Available: https://doi.org/10.1038/s41467-020-17907-w.
About This Article
Peer review information: Primary Handling Editor: Adam Khan-Qureshi. Youth STEM Matters thanks Dr Robert Spooner and Dr Simon Pitchford for their contribution to the peer review of this work.
Received: 6 May 2021
Accepted: 22 August 2021
Published: 1 October 2021