Utilizing HPLC to Analyze the Presence of Anticancerous Compounds Residing from the Isolate FM1005 (Xylaria sp.) Derived from Sinularia densa
Lela DeVine1
1Daniel K. Inouye College of Pharmacy- University of Hawaii at Hilo | Waiakea High School
Youth STEM Matters, no. 1 (2020); https://doi.org/10.51892/ysm.1.202001L. DeVine, "Utilizing High-Performance Liquid Chromatography to Analyze the Presence of Anticancerous
Compounds Residing from the Isolate FM1005 (Xylaria sp.) Derived from Sinularia densa," Youth STEM Matters,
no. 1, 2020.
If you cite this article or use it in any academic activities please let us know by e-mailing us at editor@youthstem2030.org.
Abstract
Introduction
The rapidly evolving knowledge behind the amount of natural product-based drugs that are produced by microbes is considered an area of research that should be expanded significantly [1]. The vastness of the environment provides the need for a crucial balance of environmental utilization and awareness, which may be the driving force behind potential novel compound discovery. Although natural products from marine organisms have been proposed as “a new wave of drugs” due to their great potential for supplying novel compounds for drug discovery, the attention contributed towards natural product derived drugs has been decreasing over the past nine years [1, 2].
Since the peak of natural product discovery and research in the past decade was in 2010, researchers have continued to explore marine-based medicinal studies and the isolation of microorganisms from marine sources [2]. The marine environment holds many interesting components that could lead to drug discovery in many areas, including those that involve the use of anticancer, antibacterial, and antifungal properties.
Problems may be faced when investigating the origins of these properties, as the diversity in coral reefs can make pinpointing specific sources of natural products that hold medicinal potential difficult. This is due to a general lack of understanding in the approach needed to find specific marine sources with a high compound yield within these vast areas. Additionally, there are a variety of environmental issues present in Hawaiian reefs that may negatively impact the health of marine-based natural product sources. Predominantly, sponges and corals have suffered from effects, such as bioerosion, as a result of rising ocean acidification levels. Increases in sea surface temperature have been observed since 2014 (Fig. 1A) and have led to mass bleaching events in 2015 and 2017, which damaged approximately 50% of coral in certain parts of Hawaii [3]. This resulted in declining health, reef diversity, and marine source availability issues that counteract health and pharmaceutical-based investigations.
Further issues are highlighted in Fig. 1B, in which the acidification of the water in Hawaii began to increase from January 1988. A study completed by Wisshak et al. that explored the effects of ocean acidification on sponge communities found that they are unable to regulate for changes in acidity and may be particularly vulnerable to alterations in environmental conditions, resulting in reef bioerosion [6]. Pinpointing natural product sources from reefs for potential pharmaceutical use is crucial as the marine life around us is degrading. Environmental issues, such as rising sea surface temperatures and pH increases, may hinder our ability to use the ocean as a unique medical resource for research and drug discovery in the future. Sourcing these areas first-hand, along with creating a connection between environmental utilization and awareness as to where these areas are and how vulnerable they are to marine issues, will aid in targeting uses and areas for future marine-based pharmaceutical studies.
Figure 1 – Trends in the marine environment. (A) Sea Surface Temperatures. Depictions of impacts on the marine environment derived from NOAA Coral Reef Watch, including rising sea surface temperatures from 2011 to 2016 where a peak in temperature occurred, these rises hold significant impacts on the health of marine life, especially those in shallower waters that experience these implications first. (B) Declines in pH measured at the Aloha Station. The beginning of a decreasing trend in pH levels of the waters in Hawaii, which dropped below 8.08 in January 2010. These indicate increasing impacts on marine species and habitats, and even on water quality in significantly affected areas. Figures reprinted with permission from Ref [4, 5].
Endophytic Fungi - The Source of Derived Compounds
Fungal endophytes are microorganisms that internally infect living tissues in plants and similar organisms asymptomatically. They live in a mutualistic relationship with various organisms for at least a part of their life cycle. Secondary metabolites produced by endophytes contain anticancerous, antimicrobial, antiviral, and antibacterial properties, which are used largely for commercial exploitation in the pharmaceutical and medical fields [7]. Furthermore, the chemical potential of endophytic fungi for drug discovery is still being researched.
Currently, the world population is suffering from drug-resistant microbes, such as bacteria, parasitic protozoans, and fungus, which decrease the ability of synthetic drugs to effectively work against them. An intensive search for more efficient and effective antibiotic treatments is becoming an emerging research area. Endophytic fungi establish stable symbiotic associations with their plant hosts without causing immediate symptoms or overt effects [7]. A single host species can contain many different species of endophytic fungal populations. Endophytic fungi synthesize a wide range of natural products, such as antibiotics, which have antagonistic activity against several pathogens and are commercially used for pharmaceutical, medical and agricultural purposes. Due to the benefits of their secondary metabolites, these organisms could be a great source for new drug discovery. This type of fungi can be preserved indefinitely, ensuring the continued availability of the organism. Furthermore, fungal cultures can be scaled-up to provide an inexhaustible supply of the desired metabolites and produce different bioactive compounds when culture conditions are altered [7].
Natural Products
Natural products are molecules utilized in pharmacology that are derived from organisms, including primary and secondary metabolites. Secondary metabolites are compounds predominantly derived from fungi, bacteria, and plants; they are unique and vary by source. They present a variety of biological activities and structures and are isolable in small quantities. As of 2013, several drugs derived from marine natural products are in clinical use [2]. During drug development, compound supply is one of the most challenging problems to overcome, and, as these drugs originate from marine invertebrates (previously sampled), cultivation is difficult [8]. Therefore, deriving natural products from scaled and inexhaustible cultures of endophytes may be a solution to this prominent issue.
Discovery of Novel Compounds
Studies from the Daniel K. Inouye College of Pharmacy observed novel compound discoveries from similar types of endophytic fungal strains, including Tryptoquivalines W and X, two new compounds from a Hawaiian fungal strain and their biological activities, in addition to, Circumdatin M, a new benzodiazepine alkaloid with a unique pyrimidone-4-pyrone moiety from a Hawaiian marine fungus Aspergillus 1. However, the methods in these studies differ from this current strand of research, specifically through the sample collection process and environmental links. There have not been many studies involving marine off-shore collection sites and the utilization of the Hawaiian environment, suggesting further possibilities for novel compound findings, and new approaches to understanding marine-based sources.
Previous research conducted by the author set out to find and pinpoint various species of corals and sponges, and their habitat within Hawaii's reefs in efforts to boost natural product discoveries through the isolation of endophytic fungi. This was completed to formulate a proposal as to how the scientific community can go about natural product analysis in our reefs as effectively as possible, despite their declining state. The marine samples, from which the endophytic fungal strains were isolated, were collected from Richardson Beach Park, Leleiwi Beach Park, and Puhi Bay in the town of Hilo, Hawaii. Using comparative morphological analysis, the probable identifications of the 23 isolated strains were determined, with 22% of the strains originating from Clathrina clathrus. It was concluded that sponges are the most effective and at-risk type of organism that should be prioritized for analyzation. Not only are they at risk due to rising acidity levels, but sponges tend to thrive in rockier habitats and shallower waters that contain plentiful marine life, these habitats are the first to experience any temperature changes. Strains derived from softer-based organisms were those focused upon in the research from 2019. This current research involves analyzing potentially anticancerous strains, their derived compounds, and their properties through running further antiproliferative analysis against several other cancer cell lines with compound fractions and pointing to probable identifications of these compounds.
A2780S (Ovarian Cancer), HepG2 (Liver Cancer), DU-145 (Prostate Cancer), and MCF-10A (Breast Cancer)
Based on projections from the World Health Organization, cancer-related deaths are predicted to rise to 11.4 million individuals in 2030 alone [9]. The four cancer cell lines that were involved in experimentation were prominent cancers that men and women are diagnosed with each year (refer to Appendix B for information on each cancer cell line used). According to the American Society of Clinical Oncology, in the United States alone, each year approximately 268,600 women are diagnosed with breast cancer, 174,650 men are diagnosed with prostate cancer, 33,000 individuals are diagnosed with liver cancer (of those 27,000 die as a result), and 21,000 women are diagnosed with ovarian cancer [9]. With these devastating figures, utilizing and targeting a diverse environment with sources of natural products may be vital in working towards the creation of a pharmaceutical drug that shows prominent antiproliferative activity.
Using HPLC for Compound Quantification
High-performance liquid chromatography (HPLC) is an analytical technique used to separate, identify, or quantify compounds in a subfraction through methods of column chromatography [10]. Each peak in a chromatogram indicates the presence of a compound found in the subfraction. Retention time refers to the interval of time between sample injection and peak maximum. A preparative column is representative of the initial run through the HPLC, in which the presence of compounds at certain retention times can be visualized. HPLC analysis in this project involves analyzing UV absorption rates in each isolated compound [10]. A UV visible HPLC detector uses light to analyze samples and measures the sample's absorption of light at different wavelengths. Many organic compounds of similar types and classifications fall under similar UV absorption rates and absorb UV light at specific wavelengths.
The Reversed-Phase HPLC Method
Approximately 75% of all HPLC methods use reversed-phase chromatography. Most of these protocols use an aqueous mixture of water with a miscible, polar, organic solvent, such as acetonitrile or methanol, as the mobile phase: it ensures that analytes (samples) interact properly with the non-polar, hydrophobic particle surface [11]. In the reversed-phase HPLC method, the most strongly retained compounds are non-polar, as attraction to the non-polar stationary phase is greatest. The polar compounds, being weakly retained, move the fastest through the bed towards the polar, aqueous mobile phase [11].
Research Aims
Initial goals for this research included sending ideal and potential isolated endophytic fungal strains to a local cancer center for antiproliferative assays, testing against A2780S (Ovarian Cancer), HepG2 (Liver Cancer), DU-145 (Prostate Cancer), and MCF-10A (Breast Cancer), and to gain insights into biologically active compounds that could be used for potential pharmaceuticals. To do this, a variety of methods were used, including comparing the UV absorbance rates of various compounds present in each subfraction in order to derive a probable identification of the substance.
Methods
Figure 2 - Map of Hawaii Island, USA, with general locations of targeted sample collection areas in the Hilo area. Locations were inclusive of Leleiwi Beach Park, Richardsons Beach Park, and Puhi Bay. All samples were collected with a permit from Hawaii DLNR (Department of Land and Natural Resources). Image created from GPS mapping system software.
Dive-Based Sample Collection Conducted in 2019
The criteria for collection sites varied slightly depending on the type of organism that was being collected (Fig. 2). Dives consisted of obtaining harder coral samples residing at deeper depths (~ 60ft), along with different types of sediment. Thorough investigations for smaller soft corals, sponges, and other organisms in targeted areas (small openings, cracks, and crevices present in large coral polyps and large structures of rock) took place at shallower depths. These are ideal growing areas for organisms such as these, which thrive in darker covered areas with an overhang, possibly for protection and shelter. Using hand removal techniques, amounts of Lobe coral (5cmx5cmx3cm) and other hard corals (cauliflower and finger) were measured and collected. Ideal areas for small regulated amounts of sponges and soft corals around 3cmx3cmx5cm or less were collected along large shallow rock structures.
Sample Collection Criteria and Sample Sites
Samples were collected at Leleiwi Beach Park, Richardson’s Beach Park, and Puhi Bay. The types of organisms collected were dependent upon the physical characteristics and depth of the site, with softer based organisms residing at a shallower depth, and hard corals being collected in deeper areas (refer to Appendix C for supporting details, Table C1). A variety of initial strain isolation methods were used to derive endophytic fungal strains for compound analysis, involving surface sterilization, growing the strains in media, and separation according to morphology (refer to Appendix C, Fig. C1 for more details).
Antiproliferative Analysis Methods
Antiproliferative analysis entailed sending in 10 endophytic fungal strains to a local cancer center for testing against four cancer cell lines: A2780S (Ovarian Cancer), HepG2 (Liver Cancer), DU-145 (Prostate Cancer), and MCF-10A (Breast Cancer). This was done under a testing concentration of 20μg in which the total assay time was approximately 48-72 hours. The allotted time for initial cell line growth was 24 hours, after which the cells were treated and monitored for the next 24-48-hour period. The percent viability and cytotoxic activities were further monitored and analyzed. The strains that reflected the highest antiproliferative abilities in this time were utilized for compound analysis.
Targeted Strain Culture
Solid Media Preparation and Inoculation
The isolated strains FM1005 and FM1006 were stored in a -80°C freezer. FM1005 (derived from Leather Coral), and FM1006 (derived from the Clathrina Sponge) showed the most antiproliferative action against A2780S (Ovarian Cancer), and DU-145 (Prostate Cancer) cell lines. The fungal strains were inoculated onto PDA media. The samples were inoculated onto 4 different plates and placed in an incubator at 28°C for approximately 6-7 days. Growth was monitored regularly within this period to ensure stable conditions and no contamination (refer to Appendix A for additional details).
Liquid Media Preparation and Inoculation
Approximately 250-300mL of liquid media was transferred into flasks to give the largest surface area for mycelia layer formation and was then autoclaved for an hour. Once the media had cooled, the cultivated fungal strains in the solid media were cut into small pieces, transferred into the flasks, covered, and cultured for 28 days. The focus of this portion of research is on the strain FM1005 (refer to Appendix A for additional details).
Post Fungal Culture - Preparation for Compound Extraction
To prepare for compound extraction in different gradients, the mycelia and the liquid media were filtered into large flasks. The liquid media was set aside while the mycelia were cut into small pieces and mixed with residual liquid media. An acetone fraction was created, 400mL of acetone was added to 500mL of mycelia and placed in a Sonicator overnight to break down cells. The mycelia were taken through a second filtration process to separate them from the acetone, any remaining acetone was evaporated using a rotary evaporator at 55°C. The broken-down compounds were combined with the rest of the liquid media and run through an open column gradient to extract these compounds.
Compound Extraction
When running a gradient through an open column for compound extraction, HP20 silica beads were used to remove the non-polar particle from the solution. The new media mixture was combined with the HP20 beads and evenly divided into six flasks. Once completed, the compound, in fractions of 30%, 60%, 90%, and 100%, was run through a rotary evaporator until a pure compound extract was formed (refer to Appendix A for additional details).
HLPC Analysis
Initial Runs
The initial runs of the HPLC column involved running a 40%-80% MeOH concentration for 25 minutes to act as a preparative (prep) column and lay out a baseline of compound quantification from what was first detected within this period (Fig. 3). By using the 90% compound fraction, the sample was placed in the HPLC and run to detect the presence of non-polar retained compounds from the reversed-phase method, which covers a wide range of absorption capabilities. The isolation of these peaks through running a variety of different methods allows for the identification of the UV absorption rates in each identifiable subfraction, enabling further analysis.
Figure 3 - Initial Prep Column, 40%-80% concentration for 25 minutes. Retention time is plotted against milli absorbance units (mAU) to show the initial detection of compounds within the main 90% fraction, allowing select peaks to be targeted for analysis.
Compound Separation
To isolate individual peaks for compound identification processes, a process of 40% isocratic concentration was run for 20 minutes (Fig. 4A), 70% isocratic concentration was run for 20 minutes (Fig. 4B), and a 50% isocratic concentration was run for 30 minutes (Fig. 4C). Given that peaks are representative of compound presence within a mixture, these initial runs within the subfractions show general quantification of peaks at specific retention times. Through running the HPLC at different periods of time, a methanol gradient (of varying percentages) was created, and it runs through the column containing the compound fractions (previously extracted from the open column), separating the peaks on the chromatogram until they were distinguishable and predominantly nonpolar.
Figure 4 - Methods for Compound Quantification. (A) 40% isocratic 20min (B) 70% isocratic 20min (C) 50% isocratic 30min.
Fractional Compound Analysis
HPLC analysis of further subfractions was completed by targeting isolated peaks of certain compound separation chromatographs. The probable identifications of compounds present within the fractions of FM1005 were found using a UV visible HPLC detector and Database of Absorption and Fluorescence Spectra of >300 Common Compounds for use in PhotochemCAD by Taniguchi and Lindsey [12]. In addition to deriving a probable identification, the analysis of UV absorption rates allows for both probable compound analysis and the discovery of the properties of biological macromolecules. Absorbance measurements were utilized to detect ligand binding and conformational changes, to measure concentrations, and to follow enzyme reactions. By linking this information to the HPLC analysis data, a wider range of insights into the activity, type, and structure of the compound was formed. Properties were identified using Biological Macromolecules: UV-visible Spectrophotometry by Schmid [13].
Results
Morphological Analysis and Species Origination of Isolated Strains
Figure 5 - Species origins of the 23 isolated strains. All strains prominently reside with the Clathrina Sponge, collected at both Richardson’s and Puhi Bay.
From the marine source-based strain isolation conducted in 2019, which was used for preliminary targeting, it was found that the Clathrina sponge, Haliclona sponge, and Leather coral contain higher strain isolation yields (Fig. 5). It is apparent that softer based organisms have higher strain isolation potential (Fig. 5, Table 1). Probable identification through comparative morphological analysis allowed for the general identification of Xylaria sp. in most of the marine species (Fig. 6, Table 1). Clathrina clathrus, with a 22% strain isolation yield, was collected from rocky habitats upon the underside of overhanging live rock and coral structures ranging from approximately 10-35 feet. This suggests that species from regions such as these will contain a higher number of isolable compounds upon analysis, due to increased strain yield.
Table 1 - Probable identification of the isolated fungal strains. Strains are mainly identified as Xylaria, Cerrena, and Trichoderma from morphological analysis of images. Xylaria is known to produce prominent novel bioactive compounds medicinally important in terms of their anticancer, antibacterial, and antioxidant properties.
Figure 6 - Examples of isolated endophytic fungal strains. FM1005, FM1006, FM1007, FM1009 were isolated and used in the morphological analysis process.
Antiproliferative Assay Results
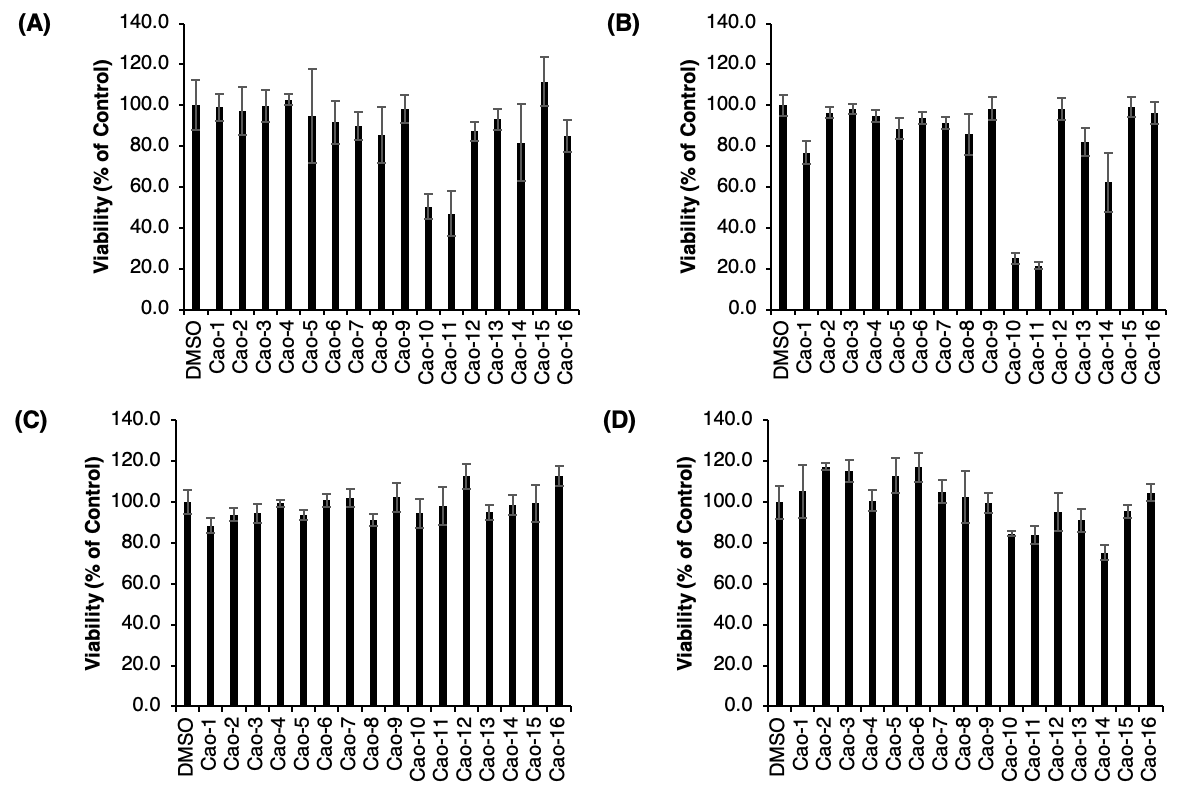
Strains 10 and 11 showed the most prominent antiproliferative action against both prostate and ovarian cancer cell lines. The percentage viability of growth decreased by approximately 50% to reach the 50% range (Fig. 7A), as well as 80% to the 20% range (Fig. 7B) in both cell lines, respectively, along with a smaller degree of decrease in viability towards breast cancer (Fig. 7D). In addition, strain 14 also showed a degree of antiproliferative action against these three cancer cell lines (Fig. 7A, B, and D).
Figure 7 - Antiproliferative Assay. The viability (% of control) after the assay was conducted is shown for each of the samples tested (1-16). (A) DU-145 (Prostate Cancer Cell Line). (B) A2780S (Ovarian Cancer Cell Line). (C) HepG2 (Liver Cancer Cell Line). (D) MCF-10A (Breast Cancer Cell Line).
Of these key data points, strains 10 and 11, which were derived from Clathrina clathrus and Sinularia densa, show similar ranges of activity against A2780S (ovarian) and DU-145 (prostate) cancer cell lines (Fig. 8A and B). One common trend supported by these data involves the marine source of the endophytes, through which the softer-based organisms show higher antiproliferative activity against the chosen cell lines compared to that of other species found in different habitats (Table 2). This indicates that within softer marine organisms, there is a correlation between higher strain isolation yields and effective antiproliferation abilities. These marine sources (predominantly sponges and soft corals) are in areas susceptible to harmful marine trends, such as changing sea surface temperatures and ocean acidification, yet they seem to be the ones that have a higher range of pharmaceutical potential. Based on overall culture conditions, to enable higher compound yield, strain 10 was chosen for further analysis.
Figure 8 - Antiproliferative Assay. The viability (% of control) after the assay was conducted is shown for each of the samples tested (1-16). (A) DU-145 (Prostate Cancer Cell Line). (B) A2780S (Ovarian Cancer Cell Line). (C) HepG2 (Liver Cancer Cell Line). (D) MCF-10A (Breast Cancer Cell Line).
Table 2 - Strains and their marine source utilized for the antiproliferative analysis. Highlighted strains are representative of the ones that exhibit the most antiproliferative action.
Analysis of Individual Subfractions
Of the 30 subfractions that were acquired throughout, those that achieved the most prominent results upon HPLC analysis were present in subfractions 10, 11, 13, 15, 18, 19, 20, 21, 23 and 25. The 90% main fraction was analyzed first because reversed-phase HPLC isolates predominantly non-polar molecules. Useable isolated molecules were most commonly found in the 90% main fraction. The most prominent range of compounds and absorption rates resided within subfraction 23 (a single set of data was used as an example, see Fig. 9 and 10). This data aids the provision of insights into which compounds within the subfraction should be focused on in further research. Within subfraction 23, the most intense peaks (1, 2, and 3) measure 13.563 mAU, 17.623 mAU, and 19.787 mAU, respectively (Fig. 9A). The molecular weight of the prominent peak of the selected portion is shown as 381.1000 m/z (Fig. 9B). Peak intensity is shown at an acquisition time range of 19-20 between tubes 5-6 (Fig. 9C), represented by counts vs acquisition time (min) allowing for better visualization of prominent compound detections from the HPLC runs. This allows for specific compound targeting for potential antiproliferative mechanisms that can be used in future studies.
Figure 9 - Subfraction 23 Analysis. (A) Baseline Chromatogram. Retention time is plotted against milli absorbance units (mAU) to show the initial detection of compounds within subfraction 23. (B) Molecular Weight. The molecular weight provides the total mass of the selected compound. The total molecular weight of the highlighted section of subfraction 23, is represented in mass to charge ratio (m/z) units. (C) Total Ion Chromatogram (TIC). The TIC represents the summed intensity across the range of masses being detected at varying points in the analysis, represented by counts vs acquisition time (min).
UV Absorption Analysis
UV Absorption graphs were the main form of data collected from a UV visible HPLC and were used to find probable identifications of compounds present within the fractions of FM1005. The collection of the UV absorption rates of each compound provided the most relevant preliminary data towards compound and structure determination targeting. Results collected from these runs suggest approximately three probable compounds per sample allowing for a wide range of identifications to be examined. Through these results in subfraction 23, it is found that the absorption intensities of peak #3 are 195.60 nm, 233.94nm, and 297.90nm (Fig. 10A). The absorption intensities of peak #1 are 215.16 nm, 256.62nm, and 317.22nm (Fig. 10B). Absorption intensities of peak #1 are 207.68 nm, 224.56nm, and 276.81nm (Fig. 10C). Absorption intensities of peak #2 are 210.50 nm, 249.89nm, and 323.81nm (Fig. 10D). These results preface probable identification-based analysis and are representative of each compound within endophytic fungal strain FM1005, which allows for further information to be gathered on the nature of each compound. Through the collection of this set of data, a total UV range of 195-384nm was present from 10 subfractions and 48 compounds within the 90% major fraction. This wide range of results also suggests that the marine sources are particularly useful for this type of compound-based research, due to the high compound yield within this main fraction alone.
Figure 10- UV Absorption Rates Derived from HPLC Detector. All graphs depict UV wavelength (nm), plotted against peak absorption percentage. Each peak is representative of the dominant absorption rate of the compound at differing wavelength values, selected at varying retention times. (A) Subfraction 23, T24-25. Absorption intensities of peak #3 at both 19.69 min and 19.86 min. (B) Subfraction 23, T5-6. Absorption intensities of peak #1 at both 13.44 min and 13.66 min. (C) Subfraction 23, T20-21. Absorption intensities of peak #1 at both 18.48 min and 18.66 min. (D) Subfraction 23, T17-18. Absorption intensities of peak #2 at both 17.51 min and 17.74 min.
Fractional Compound Analysis, Probable Identification Based Upon UV Absorption Rates and Molecular Weight
The probable identifications of compounds present within the fractions of FM1005 were found using Database of Absorption and Fluorescence Spectra of >300 Common Compounds for use in PhotochemCAD by Taniguchi and Lindsey [12], based upon similarities in UV Absorption Rates. Various sections of this database were analyzed to determine similar ranged compounds (see Appendix C for all identified structures derived from UV absorption rates.). The three main compounds considered common within the probable compound data set are Benzene, Rac-BINAP, and L-Histidine (see Appendix C, Fig. C2 for structures). These compounds have a UV absorption rate of 225nm, 228nm, and 206nm, respectively. Compounds that fall under similar absorption categories will tend to have similar structures and natures. All of the identified compounds from HPLC analysis were culminated to form a data set of probable compound isolations based upon UV absorption rates, the data set depicts the selected major fraction, subfraction, compound number/sample code, the molecular weight of the compound, detected UV absorption rates, as well as the finalized probable compound identification (Table 3).
Table 3 - Dataset of Probable Compounds. Details of the probable compound identifications from UV absorption data are shown for each subfraction under the 90% major fraction.
PCR Analysis of FM1005
Marine-source derived endophytic fungal strain FM1005 was sent out for PCR analysis to confirm morphological analysis efforts made in 2019 (Fig. 11). Results confirm the identification of FM1005 as Xylaria sp. isolated from Sinularia densa, or Dense Leather Coral.
Figure 11 – PCR Results. Additional details on the specific amino acid sequence derived specifically from the PCR analysis. Further information can be found under the GenBank (National Center for Biotechnology Information), under access code MT126321. Refer to Appendix A for more details on PCR background.
Discussion
This research aimed to provide compound and antiproliferative analysis of endophytic fungal strains derived from marine sources. A variety of different marine species were collected from areas of the reef through scuba diving and hand removal techniques. These samples then underwent a series of processes, through which endophytic fungal strains were derived from. Compound fractions were also extracted and put through a series of comparative morphological analysis, antiproliferative assays, HPLC, and PCR.
Within the general data from collection sites and the relationship between species origination and strain isolation yield, it was found that softer based organisms found in rockier and shallower depths have higher strain isolation potential. This statement is supported by Fig. 5, which shows that the Clathrina sponge, Haliclona sponge, and the Leather coral have the highest strain isolation yield and potential. Specifically, the species with the highest strain isolation yield of 22% was Clathrina clathrus, which is an irregular mass of anastomosed tubes tending to be mostly white and yellow with occasional brown and red tinges. This particular species of sponge was mainly collected in rocky habitats upon the underside of overhanging live rock and coral structures, which ranged from approximately 10-35 feet depending on the beach and region (shallower areas tending to be Richardson’s shallow coral population). Through conducting comparative morphological analysis, the strains isolated were speculated to be primarily Xylaria sp., Cerrena, and Trichoderma [14].
The antiproliferative analysis was conducted with four prominent cancer cell lines: A2780S (Ovarian Cancer), HepG2 (Liver Cancer), DU-145 (Prostate Cancer), and MCF-10A (Breast Cancer). Results as shown by the antiproliferative analysis (Fig. 8) also follow a similar trend, in which endophytic fungal strains FM1005 and FM1006 derived from the same marine habitat also have significant antiproliferative abilities: they mostly effect ovarian and prostate cancer cell lines, dropping their growth by 80% and 50%, respectively. These come from softer based organisms, which also hold higher strain isolation potential. The ability of these species types to hold these antiproliferative and strain isolation potentials may be due to the niche of this species in the environment - softer based organisms, such as sponges, are known to play a role in the regulation and cycling of nutrients in their environment [15]. The marine organisms that have shown to be valuable through possessing a high isolation yield are those whose habitats are especially susceptible to rising water temperatures, ocean acidification, as well as other effects of pollution. This data is crucial for the scientific community’s understanding of natural product sources. Understanding the peak conditions as to where significant sources reside holds a role in pinpointing natural product sources from reefs. The potential of prospective pharmaceuticals derived from the marine environment is rising. However, as environmental issues such as rising pH and sea surface temperatures take place, sourcing softer based organisms in shallower waters could become problematic and, therefore, prevent environmental utilization.
The inner workings and mechanisms found in the results that show antiproliferative activity with ovarian cancer (A2780S), prostate cancer (DU-145), and a degree of activity against breast cancer (MCF-10A) are not fully known, as further experimentation may need to be completed to gain a thorough understanding of this topic. However, the potential links and backgrounds of these three types of cancers, which are known to be genetically inherited, are open to discussion. Inherited genetic mutations in the BRCA2 gene, a human tumor repressor gene that is responsible for repairing DNA, is known to have links to these three cell lines. Targeted mechanisms for later experimentation that should be researched include; inhibitory mechanisms against this gene, the potential mechanism for the reactivation of apoptosis (programmed cell death) within these cell lines and alterations in differing metabolic activities.
In addition to the initial strain isolation and antiproliferative assays, four major fractions (30%, 50%, 90%, 100%), 10 subfractions under the 90% major fraction, and the UV absorbance rates of 48 compounds were derived using HPLC methods. Fractional compound analysis for the probable identification based upon UV absorption rates and molecular weight were derived for 48 compounds. The reasoning behind using this type of data comparison to derive an accurate insight revolves around UV visible HPLC detectors having the ability to use light to analyze samples and measure the sample's absorption of light at different wavelengths. Many organic compounds of similar types and classifications fall under similar UV absorption rates and absorb UV light at specific wavelengths. Therefore, this method of analysis forms accurate insights on related structures and serves a purpose in deriving proposed novel compound structures for future ongoing research. The design and nature of new molecules for photochemical studies are focused upon specific knowledge of spectral features, including the absorption spectrum, fluorescence spectrum, and fluorescence quantum yield. In cases of analyzation such as this, fluorescent properties may be of direct relevance and extremely useful as proxies to gain insights into the nature of the specific compound [12]. The probable compound identification table provides full absorption rate details of all of the isolated compounds listed in table 3. Through the works of Taniguchi and Lindsey, it was found that a collection of this data showed a prominence of compounds that have similar UV absorption rates to Benzene, Rac-BINAP, and L-Histidine, which can be looked towards when analyzing the structure, nature, and types of compounds in varying fractions. The total UV range that these compound samples fell under was 195-384 nm, with most of these samples residing in the 200 nm range (see Fig. C2).
With this understanding of the most prominent UV range that the compounds fell into, the properties of the biological macromolecules could be addressed. This allows various structural aspects that resided within the UV range of the marine-based compounds to be acknowledged for future references in compound-based research. Absorbance measurements were used to measure concentrations, to detect conformational changes and ligand binding, and to follow enzyme reactions. Linking this information to the HPLC analysis data enables a wider range of insights into the activity, type, and structure of the compound to be formed. Through analysis of the work of Schmid, the prominent range of UV absorption at 200 nm was focused upon [13]. Through this, it was found that buffers, which contain carboxyl and/or amino groups, absorb light below 220 nm; the concentrations of nucleic acids in solution are determined by their strong absorbance rates at the range of about 260 nm, and disulfide bonds that form between two cysteine residues also show an absorbance band near 260 nm. In addition to this, insights into the types of molecules in this general range point to aspects behind the absorption rates of proteins, in which proteins show absorption maxima between 275 and 280 nm, and peptide groups of the protein absorb light in a UV range from 180-230 nm [13]. For example, regarding the absorption maxima of proteins, UV absorption rates fall under the range of those similar to the probable compounds and are caused by the absorbance of the two aromatic amino acids, tryptophan (Trp) and tyrosine (Tyr). There is also potential that these structures are also caused by the absorption of cystine (i.e. from disulfide bonds) [13]. This allows for the targeting of certain structural aspects when later determining the overall structure of specific compounds. It is crucial to understand these compounds because many of them have a high potential of being novel, and an accurate proposed structure is necessary.
Marine-source derived endophytic fungal strain FM1005’s PCR analysis confirmed morphological analysis efforts made in 2019 as Xylaria sp. isolated from Sinularia densa, or Dense Leather Coral. In this portion of experimentation, PCR analysis is important as solidifying the source of these compounds will allow for further analysis of Xylaria sp. in relation and comparison to other types of fungal strains. There can also now be an accurate comparison of different results derived from Xylaria sp. and found from different studies. Through this confirmation, further insight, and research, the discussion of the importance of Xylaria sp. in the natural products based pharmaceutical sphere can be elaborated on. In a study conducted by Adnan et al., Xylaria sp. was found to produce wide classes of bioactive compounds, some of which can be used to create mixtures that inhibit the survival and cell growth against human lung cancer (carcinoma) cell line A549 [8]. These links with Xylaria sp. have been proven to show prominent anticancer and antibacterial properties. Finding links such as these through PCR confirmation provides the ability to create a solidified baseline of characteristics to analyze and connect for potential inhibition mechanisms, as mentioned earlier in previous discussion sections.
Because there is little published literature on the specifics of endophyte derived compounds from marine sources, it is difficult to pinpoint specific sources of error in these findings. Once structures are determined, there is a possibility for a variety of them to be considered novel to the scientific community. Although sources of error remain unclear, it is important to note that this portion of research only covers the compounds found within the 90% major fraction, meaning that these findings are considered preliminary and minuscule in the grand scheme of this research. Specifically, in terms of compound targeting, there are three other major fractions to be explored, as well as the entirety of FM1006, which also showed prominent antiproliferative activity. This suggests that there are many more possibilities into the discovery of an antiproliferative compound that could be targeted and used for further pharmacological research and brings light upon how broad the scope of marine endophyte derived compounds can span. Future steps that can be taken to further this research would involve running more antiproliferative analysis and potential bioassays against non-cancerous cells (HEK 293 human embryonic kidney cells), targeting specific prominent compounds for antiproliferative analysis, and conducting further antibacterial assays. It would also be highly beneficial to work with members of the lab in efforts to derive a proposed structure and finalize any novel compounds that possess potential in the pharmaceutical industry.
Through findings present in this experimentation, there is potential for innovative marine medical based applications that antiproliferative compounds within these isolated fungal strains could have the ability to contribute to. Future research will link directly with these findings and possibilities and be focused more specifically on the pharmacological aspect of this study. Xylaria sp. can be preserved indefinitely, ensuring the continued availability of leather coral species in the reef, and preventing environmental degradation. Many times, even after surgeries have been completed to remove a tumor, there is potential for it to resurface, coming at an unfortunate cost to the patient, and may even lead to death if unknown or untreated. There are instances during surgery that not all of the tumor is visible, or sometimes, it is inaccessible. A potential application of this data in terms of drug-based development would be to create a drug from targeted isolated compounds from FM1005 that could be applied to the general area of the tumor during surgery to reduce the probability of cancer resurfacing in the patient. This could be a crucial aspect in finding a safer alternative to chemotherapy. Factors such as pharmacokinetics, pharmacodynamics, and the impacts of specific compound fractions against noncancerous cells would have to be considered, as well as potential ways to administer the drug systemically through antibody targeting. Specific pharmacokinetics that need to be acknowledged if various compound fractions, such as these, were created into a potential drug would be inclusive of, varying levels of effectiveness on the site administered and the site of action, and the concentrations of the drug found in plasma and tissues sampled at varying locations. It is also crucial to look into cellular response upon administration of a potential drug with varying concentrations of these compounds, which may impact both cell and drug effectiveness. Finding a balance between these two factors will be crucial to search for in further research. This will entail the measurement of receptors on the cell surface and how this holds an impact on the creation of various signal transduction pathways that may impact gene regulation, protein production, inhibitory factors, expression of the creation of certain hormones and toxic drug complications [16].
This research, and further research like it, will continue to acknowledge a crucial balance point of environmental utilization and awareness for potential future uses in the medical field.
Acknowledgements
The author would like to thank Dr. Shugeng Cao and Mr. KH A Zaman from the Daniel K. Inouye College of Pharmacy, as well as Mrs. Whitney Aragaki and Dr. Chad Higa from Waiakea High School, Mr. Phillip Caporusso from East Hawaii Divers, and Gary and Monica DeVine for their continued support throughout the entire research process.
Author's Notes
All figures, images, or tables were created by the author, unless otherwise mentioned in the description provided for said figure.
References
[1] D. J. Newman and G. M. Cragg, “Natural Products as Sources of New Drugs from 1981 to 2014,” Journal of Natural Products, vol. 79, no. 3, pp. 629–661, 2016. Available: https://doi.org/10.1021/acs.jnatprod.5b01055.
[2] R. Monaster and H. Luesch, “‘Marine Natural Products: A New Wave of Drugs?,’” Future Medicinal Chemistry, vol. 3, no. 12, pp. 1475-1489, Sep. 2011. Available: https://doi.org/10.4155/fmc.11.118.
[3] Coral Health Atlas, “Widespread Coral Bleaching Event to Hit U.S. Hard for Third Straight Year,” Coral Health Atlas, 2018. [Online]. Available: https://hilo.hawaii.edu/coralhealth/news/widespread-bleaching.php.
[4] C. Kane, “'Big, hot blob' puts Hawaiian reefs at risk,” Conservation International, 22-Oct-2015. [Online]. Available: https://www.conservation.org/blog/-big-hot-blob-puts-hawaiian-reefs-at-risk. [Figure data derived from NOAA Coral Reef Watch].
[5] Anon, “Ocean acidification,” European Environment Agency, 24-Jun-2020. [Online]. Available: https://www.eea.europa.eu/data-and-maps/indicators/ocean-acidification-3/assessment.
[6] M. Wisshak, C. H. L. Schönberg, A. Form, and A. Freiwald, “Effects of ocean acidification and global warming on reef bioerosion—lessons from a clionaid sponge,” Aquatic Biology, vol. 19, no. 2, pp. 111–127, 2013. Available: https://doi.org/10.3354/ab00527.
[7] P. Kumar Pandey, S. Singh, R. Narayan Singh Yadav, A. Kumar Singh, and M. C. Kumar Singh, “Fungal endophytes: Promising tools for pharmaceutical science,” International Journal of Pharmaceutical Sciences Review and Research, vol. 25, no. 2, pp. 128–138, Mar. 2014.
[8] M. Adnan, M. Patel, M. N. Reddy, and E. Alshammari, “Formulation, evaluation and bioactive potential of Xylaria primorskensis terpenoid nanoparticles from its major compound xylaranic acid,” Scientific Reports, vol. 8, no. 1, pp. 1-12, Jan. 2018. Available: https://doi.org/10.1038/s41598-018-20237-z.
[9] World Health Organization, “Cancer,” World Health Organization. [Online]. Available: http://www.who.int/health-topics/cancer#tab=tab_1.
[10] T. E. Crowley, Purification and Characterization of Secondary Metabolites- A Laboratory Manual for Analytical and Structural Biochemistry, Academic Press, 10 Aug. 2019.
[11] Waters Corp., “HPLC Separation Modes,” Waters, 2020. [Online]. Available: http://www.waters.com/waters/en_US/HPLC-Separation-Modes/nav.htm?cid=10049076&locale=en_US.
[12] M. Taniguchi and J. S. Lindsey, “Database of Absorption and Fluorescence Spectra of >300 Common Compounds for Use in PhotochemCAD.,” Photochemistry and Photobiology- Wiley Online Library, vol. 94, no. 2, pp. 290-327, Nov. 2017. Available: https://doi.org/10.1111/php.12860.
[13] F. X. Schmid, “Biological Macromolecules: UV‐Visible Spectrophotometry,” Encyclopedia of Life Sciences- Wiley Online Library, Apr. 2001. Available: https://doi.org/10.1038/npg.els.0003142.
[14] N. S. Chowdhury, M. H. Sohrab, S. R. Rony, S. Sharmin, M. Begum, M. Rana, and C. Hasan, “Identification and bioactive potential of endophytic fungi from Monochoria hastata (L.) Solms,” Bangladesh Journal of Botany, vol. 45, no. 1, pp. 187-193, Mar. 2016.
[15] C. Goodwin, R. Rodolfo-Metalpa, B. Picton, and J. Hall-Spencer, “Effects of ocean acidification on sponge communities,” Marine Ecology, vol. 35, no. 1, pp. 41–49, 2014. Available: https://doi.org/10.1111/maec.12093.
[16] S. Senapati, A. K. Mahanta, S. Kumar, and P. Maiti, “Controlled Drug Delivery Vehicles for Cancer Treatment and Their Performance,” Signal Transduction and Targeted Therapy- Nature, vol. 3, no. 1, pp. 1-19 Mar. 2018. Available: https://doi.org/10.1038/s41392-017-0004-3.
Appendices
Appendices A, B and C are available in the PDF version of this article.
About This Article
Peer review information: Primary Handling Editor: Adam Khan-Qureshi. Youth STEM Matters thanks Poppy Bradley, Kerena Norris, Lucy Chen and Elizabeth Bourn for their contribution to the peer review of this work.
Received: 24 April 2020
Accepted: 28 July 2020
Published: 19 August 2020
Publisher’s note: Youth STEM 2030 remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.